Презентация на тему: Directed Mutagenesis and Protein Engineering

Directed Mutagenesis and Protein Engineering
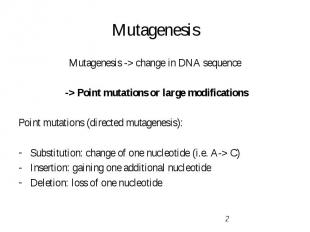
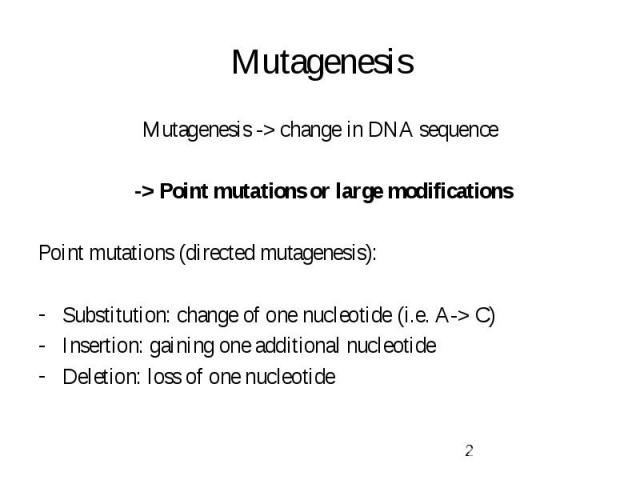
Mutagenesis Mutagenesis -> change in DNA sequence -> Point mutations or large modifications Point mutations (directed mutagenesis): Substitution: change of one nucleotide (i.e. A-> C) Insertion: gaining one additional nucleotide Deletion: loss of one nucleotide
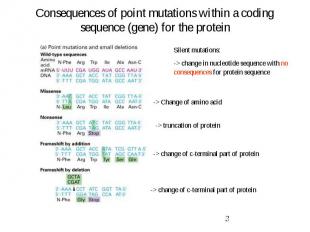
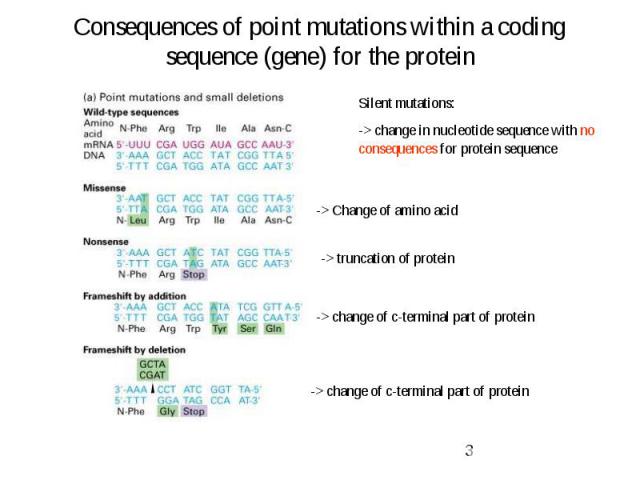
Consequences of point mutations within a coding sequence (gene) for the protein
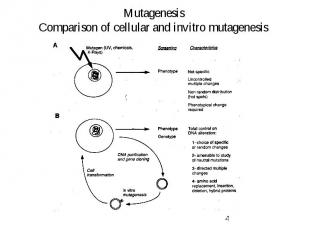
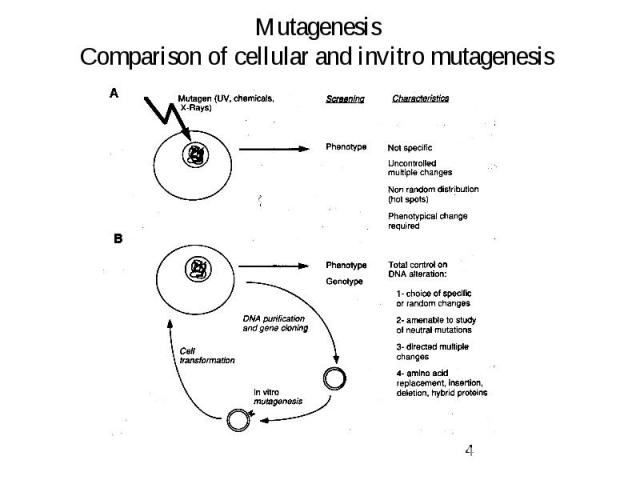
Mutagenesis Comparison of cellular and invitro mutagenesis
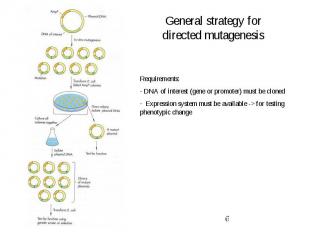
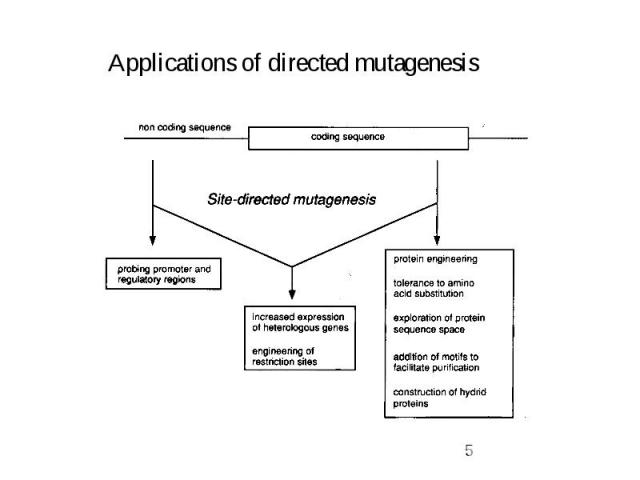
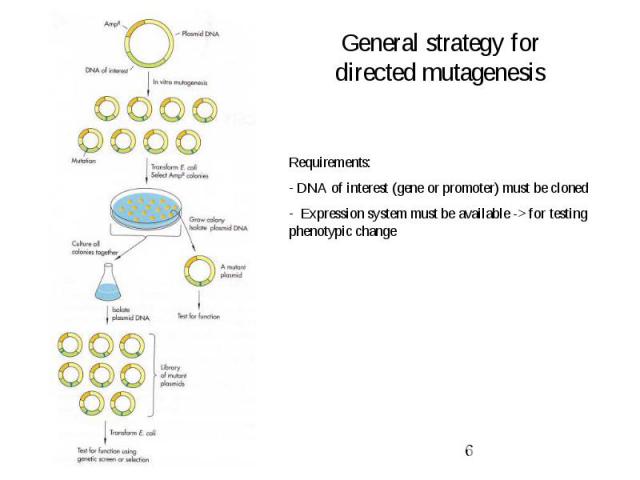
General strategy for directed mutagenesis

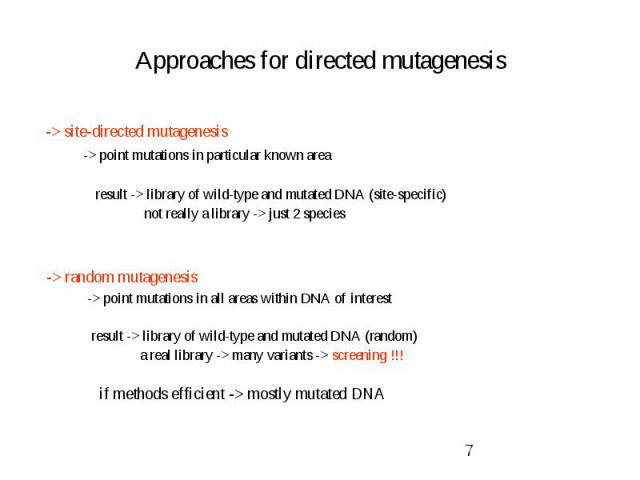
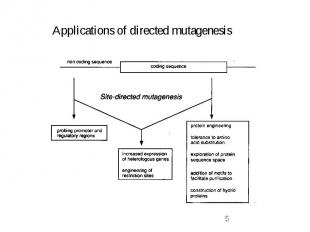
Approaches for directed mutagenesis -> site-directed mutagenesis -> point mutations in particular known area result -> library of wild-type and mutated DNA (site-specific) not really a library -> just 2 species -> random mutagenesis -> point mutations in all areas within DNA of interest result -> library of wild-type and mutated DNA (random) a real library -> many variants -> screening !!! if methods efficient -> mostly mutated DNA
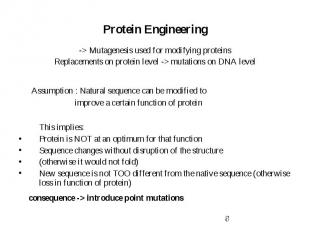
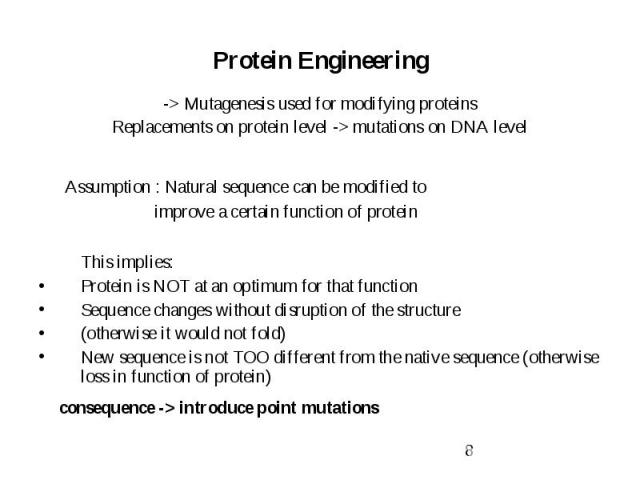
Protein Engineering -> Mutagenesis used for modifying proteins Replacements on protein level -> mutations on DNA level Assumption : Natural sequence can be modified to improve a certain function of protein This implies: Protein is NOT at an optimum for that function Sequence changes without disruption of the structure (otherwise it would not fold) New sequence is not TOO different from the native sequence (otherwise loss in function of protein) consequence -> introduce point mutations
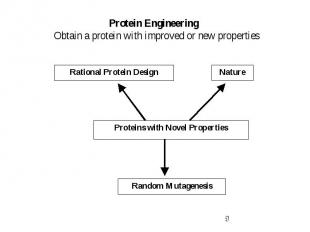
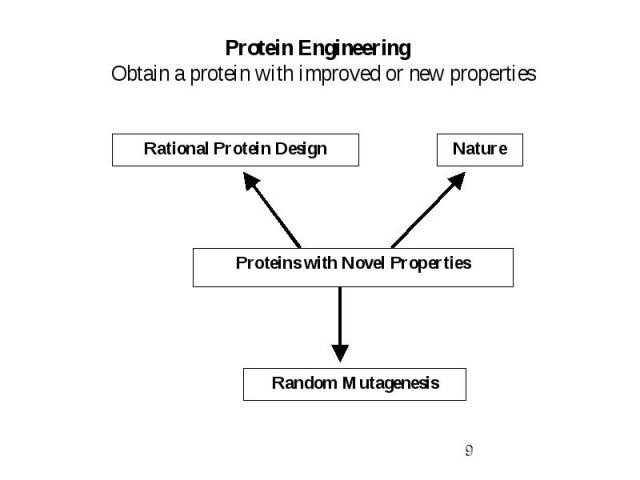
Protein Engineering Obtain a protein with improved or new properties

Rational Protein Design
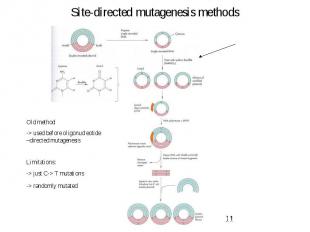
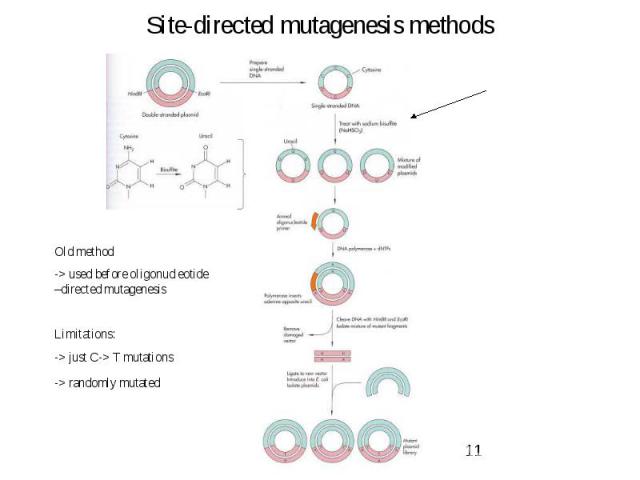
Site-directed mutagenesis methods
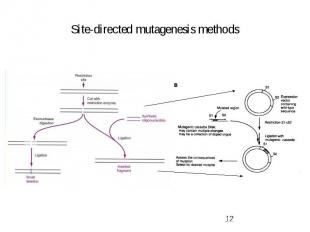
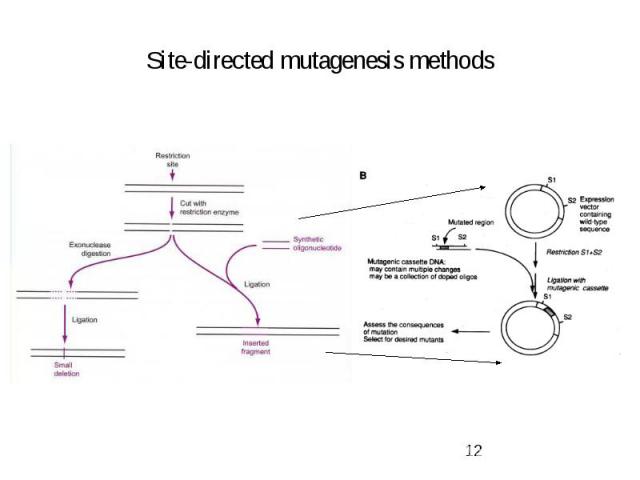
Site-directed mutagenesis methods
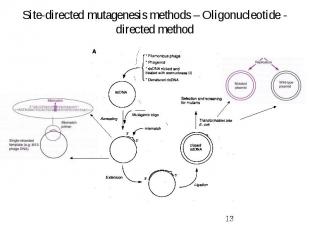
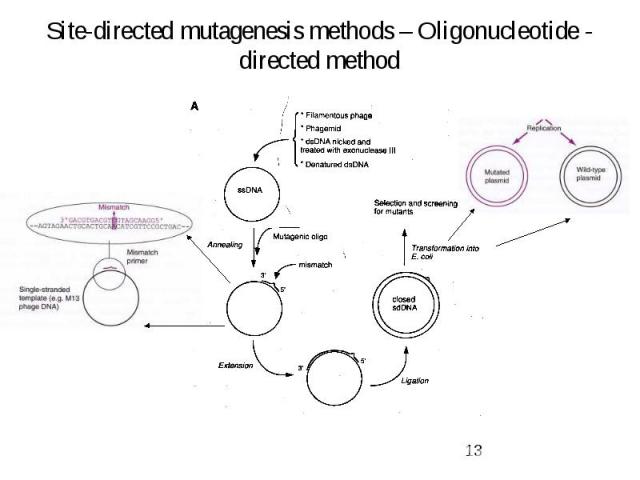
Site-directed mutagenesis methods – Oligonucleotide - directed method
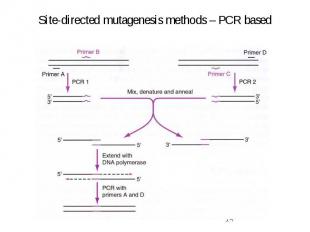
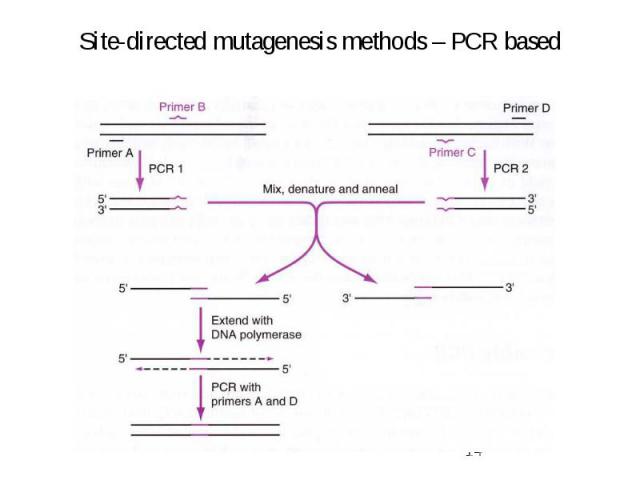
Site-directed mutagenesis methods – PCR based
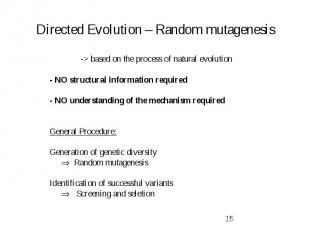
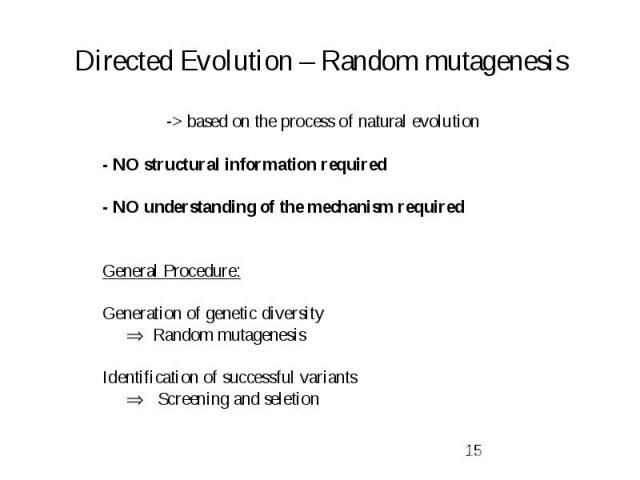
Directed Evolution – Random mutagenesis
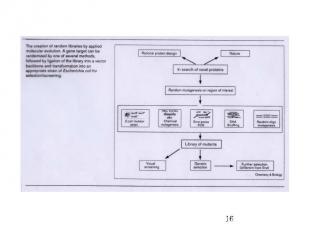
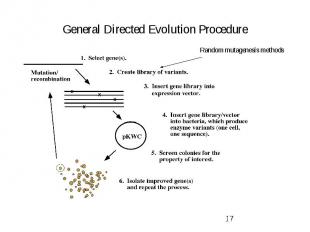
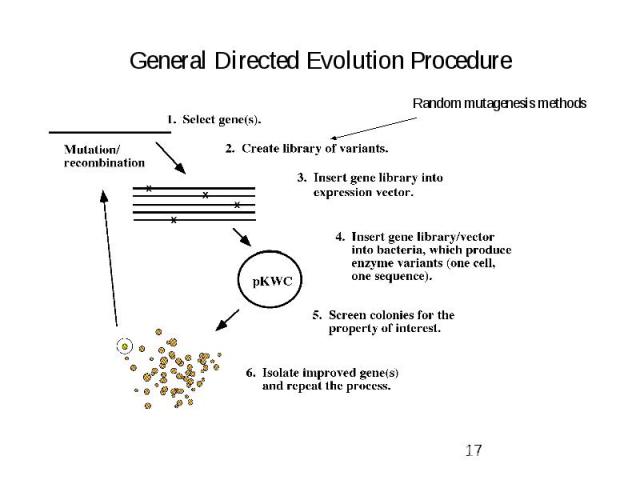
General Directed Evolution Procedure

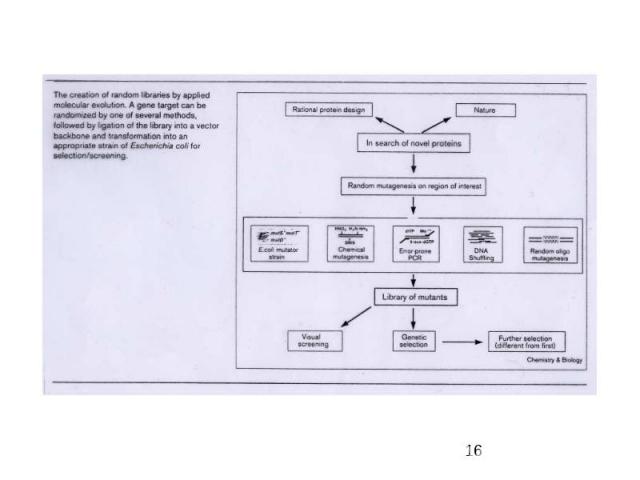
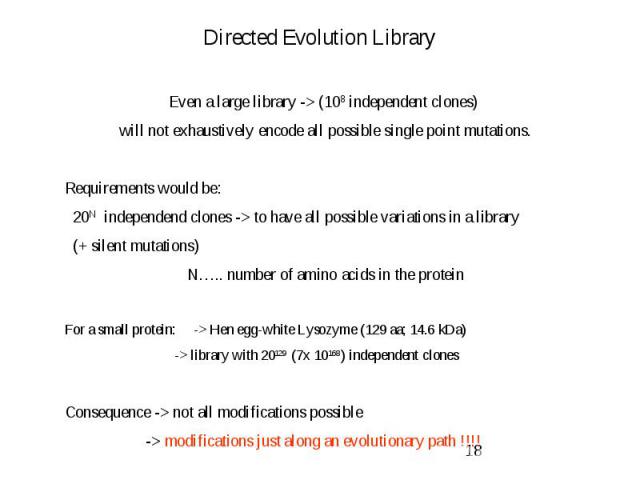
Directed Evolution Library

Limitation of Directed Evolution

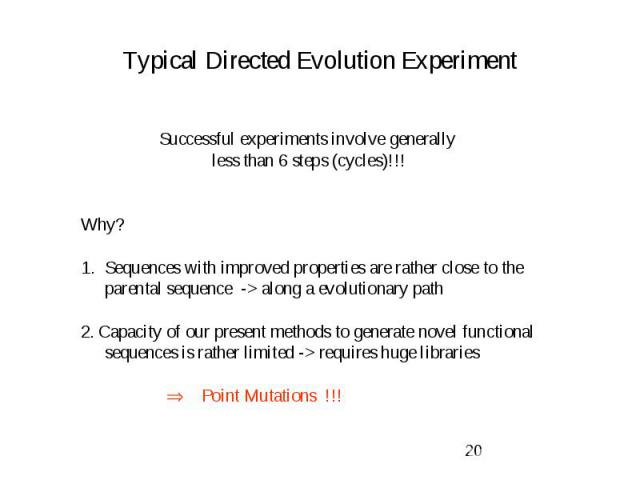
Typical Directed Evolution Experiment
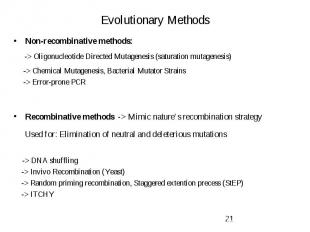
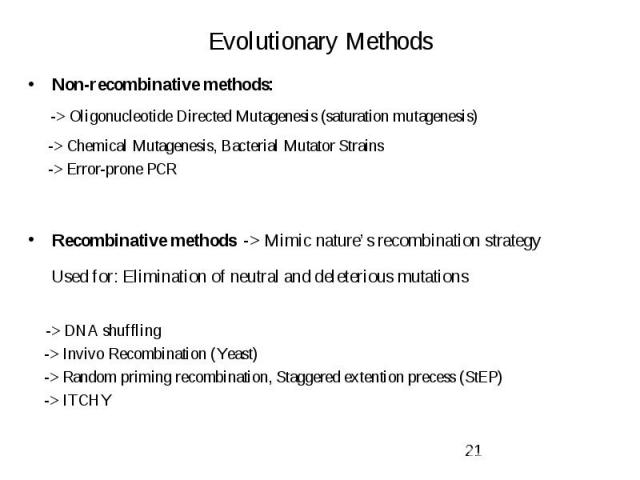
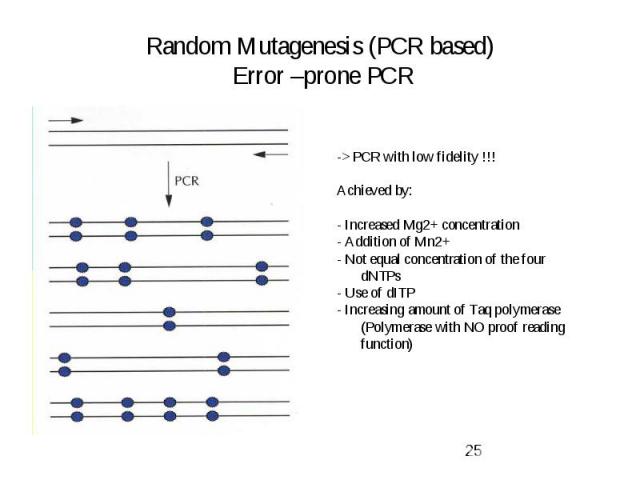
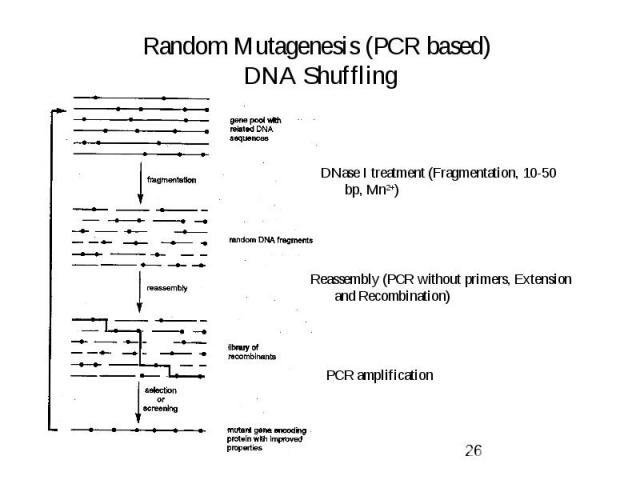
Evolutionary Methods Non-recombinative methods: -> Oligonucleotide Directed Mutagenesis (saturation mutagenesis) -> Chemical Mutagenesis, Bacterial Mutator Strains -> Error-prone PCR Recombinative methods -> Mimic nature’s recombination strategy Used for: Elimination of neutral and deleterious mutations -> DNA shuffling -> Invivo Recombination (Yeast) -> Random priming recombination, Staggered extention precess (StEP) -> ITCHY

Evolutionary Methods Type of mutation – Fitness of mutants Type of mutations: Beneficial mutations (good) Neutral mutations Deleterious mutations (bad) Beneficial mutations are diluted with neutral and deleterious ones !!! Keep the number of mutations low per cycle -> improve fitness of mutants!!!
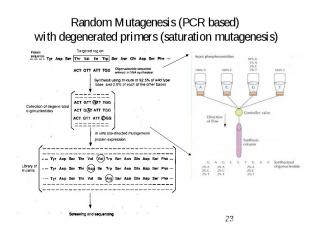
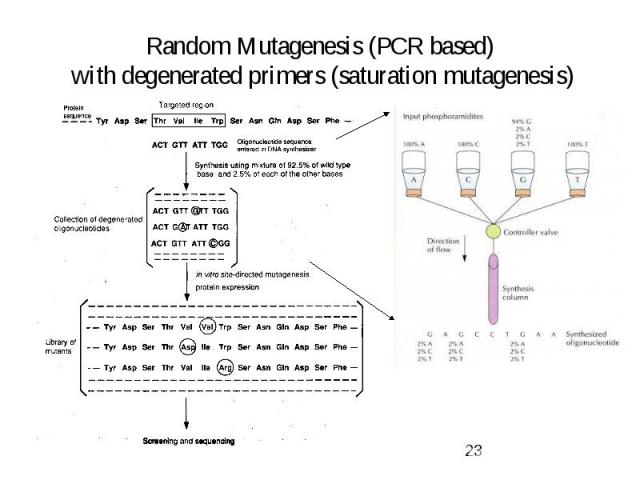
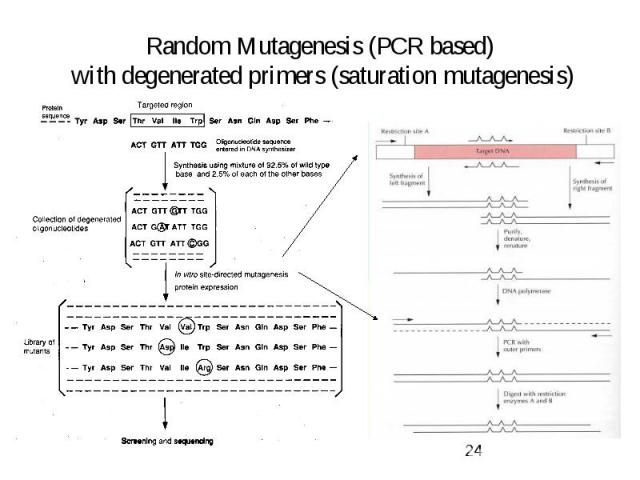
Random Mutagenesis (PCR based) with degenerated primers (saturation mutagenesis)
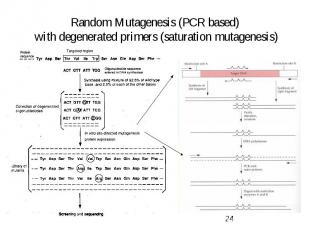
Random Mutagenesis (PCR based) with degenerated primers (saturation mutagenesis)
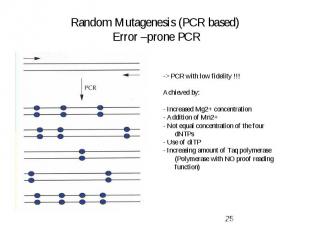
Random Mutagenesis (PCR based) Error –prone PCR
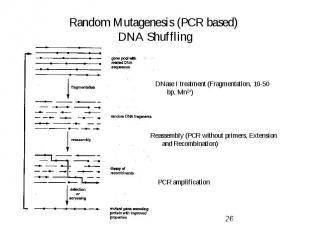
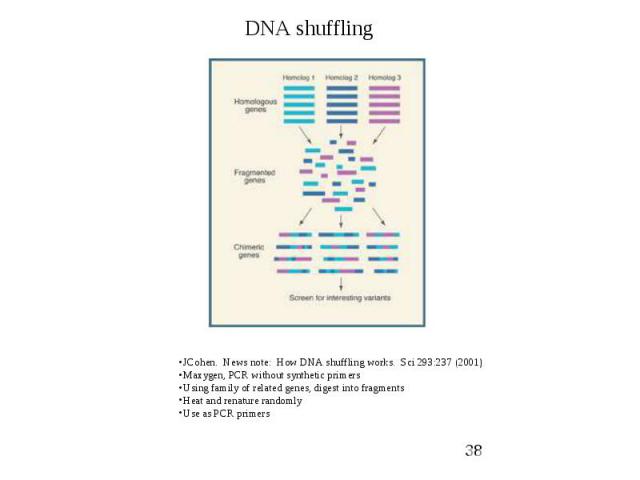
Random Mutagenesis (PCR based) DNA Shuffling
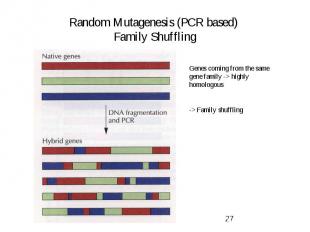
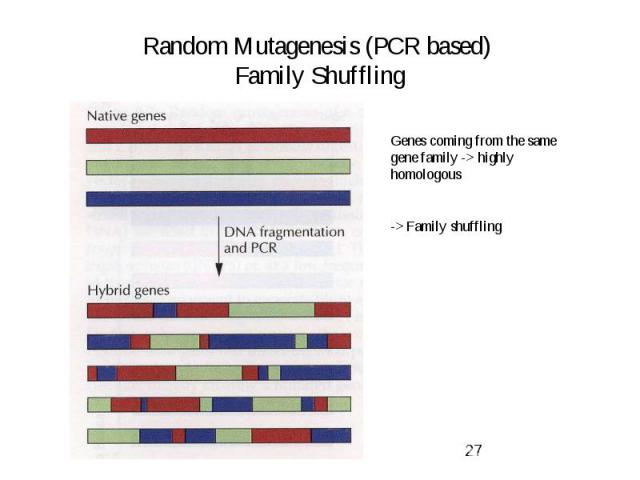
Random Mutagenesis (PCR based) Family Shuffling
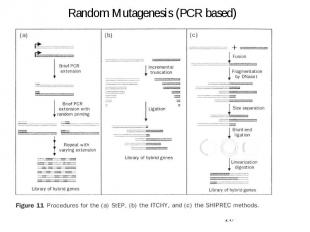
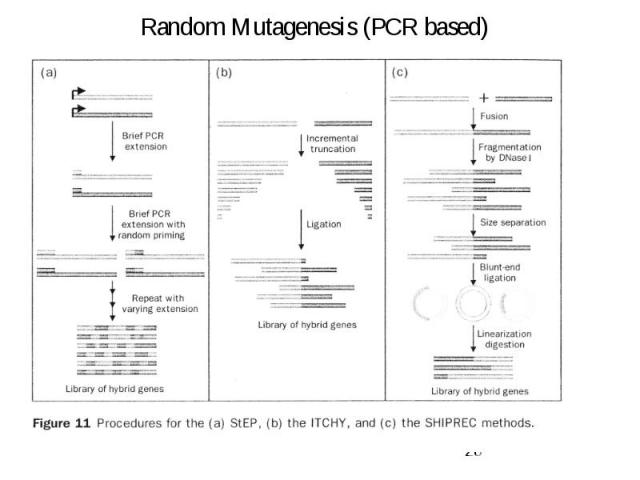
Random Mutagenesis (PCR based)
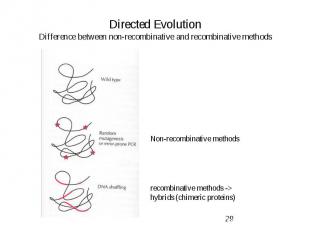
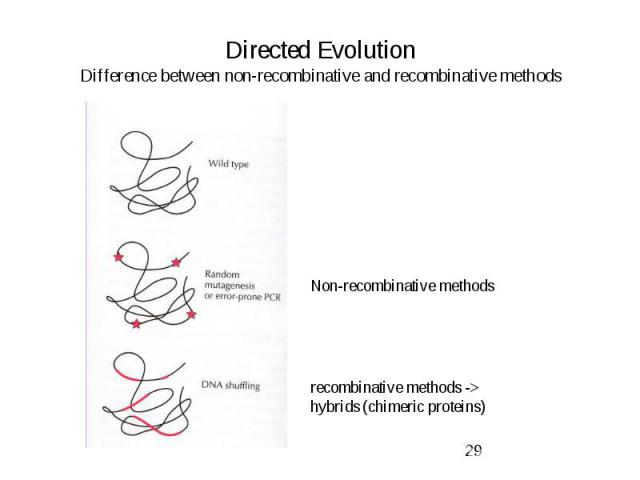
Directed Evolution Difference between non-recombinative and recombinative methods
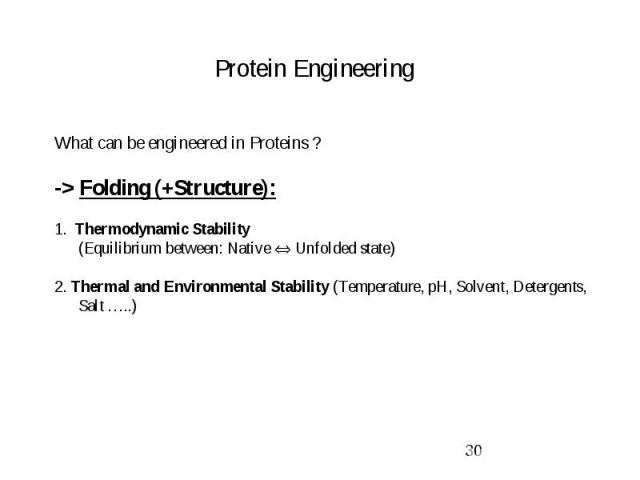
Protein Engineering
Protein Engineering

Protein Engineering
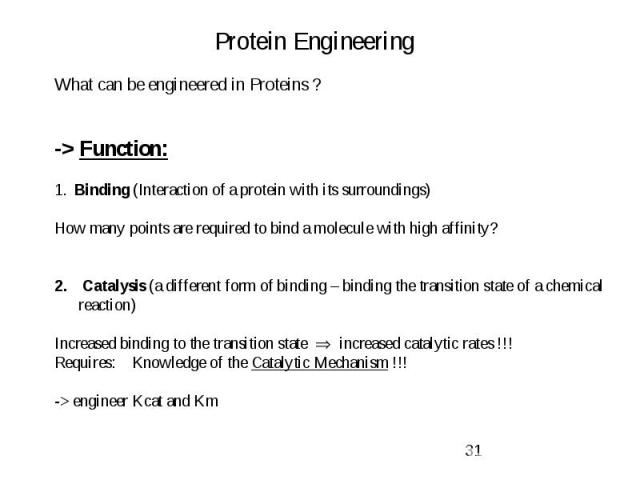
Protein Engineering
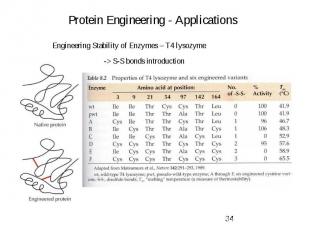
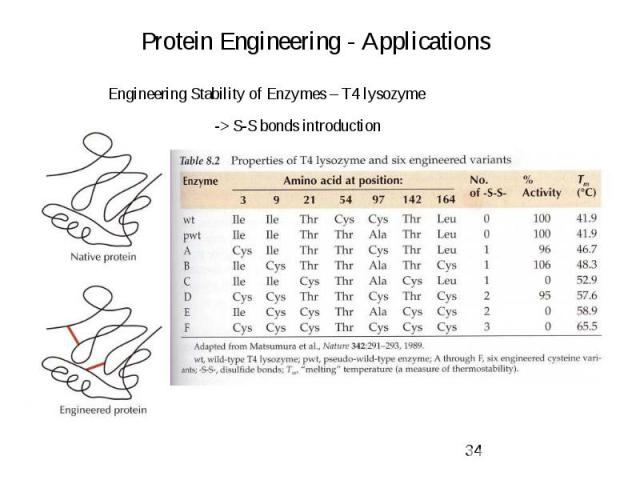
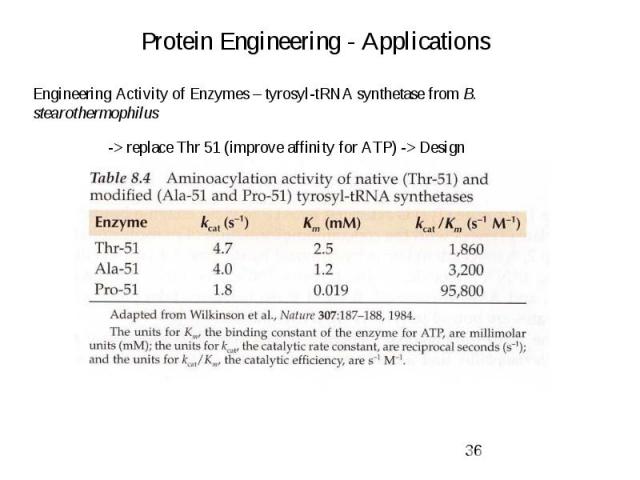
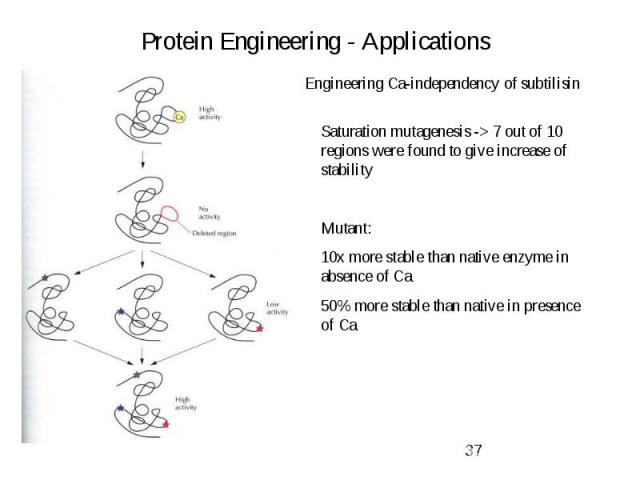
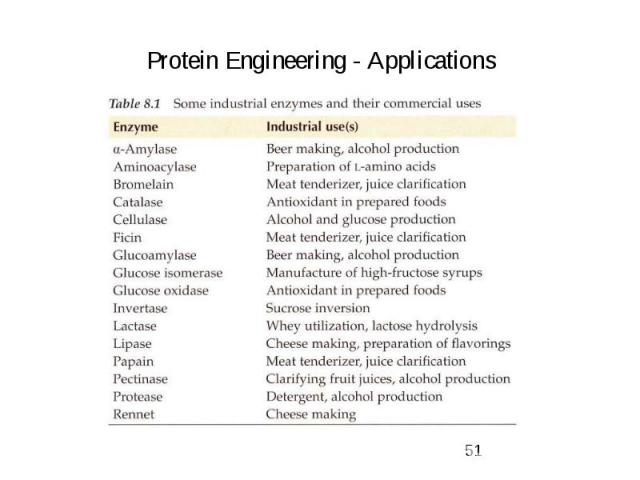
Protein Engineering - Applications
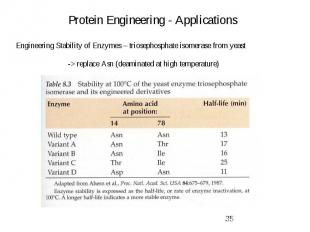
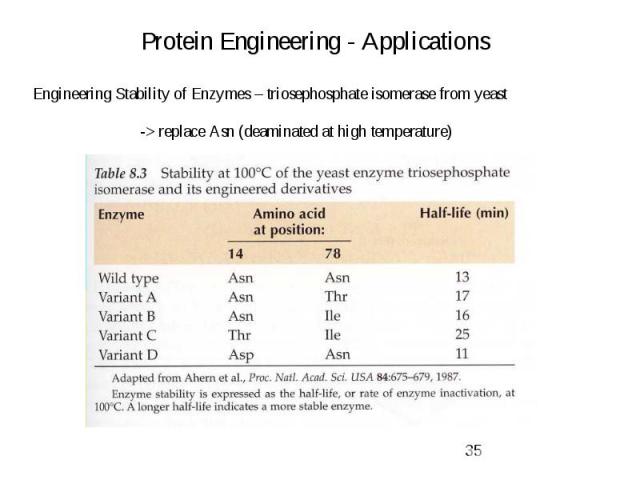
Protein Engineering - Applications
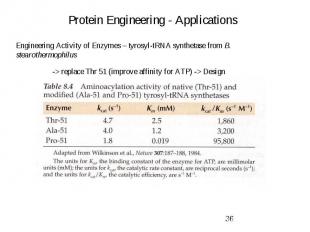
Protein Engineering - Applications
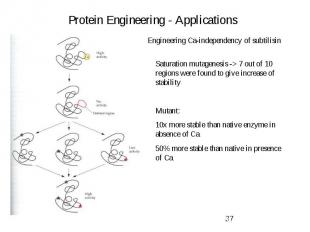
Protein Engineering - Applications
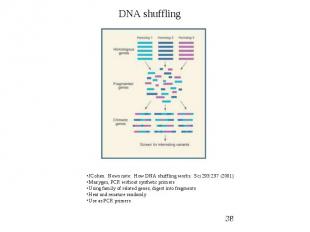
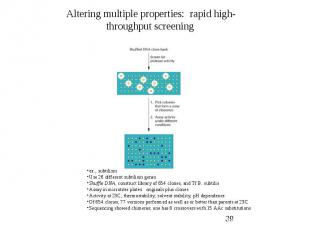


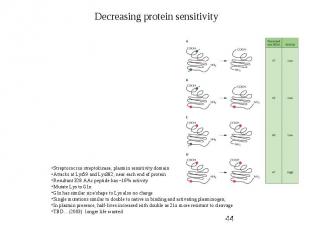
Protein Engineering - Applications
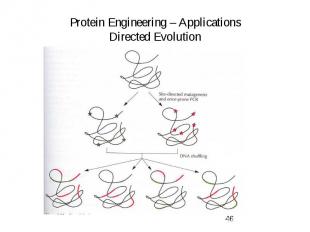
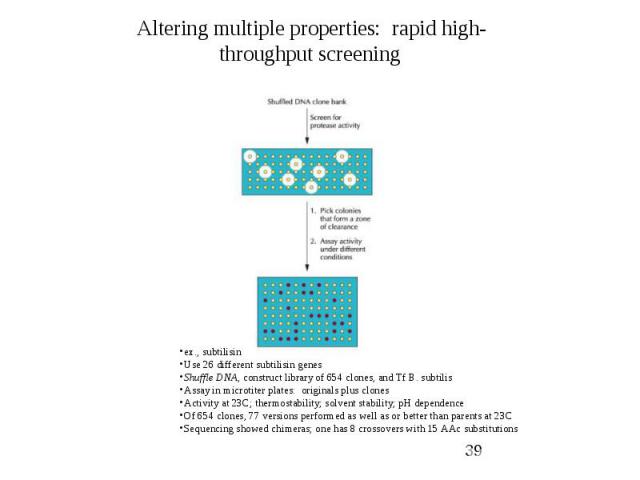
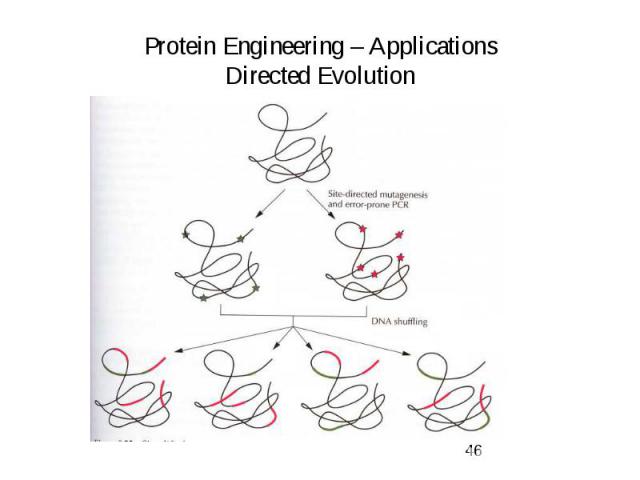
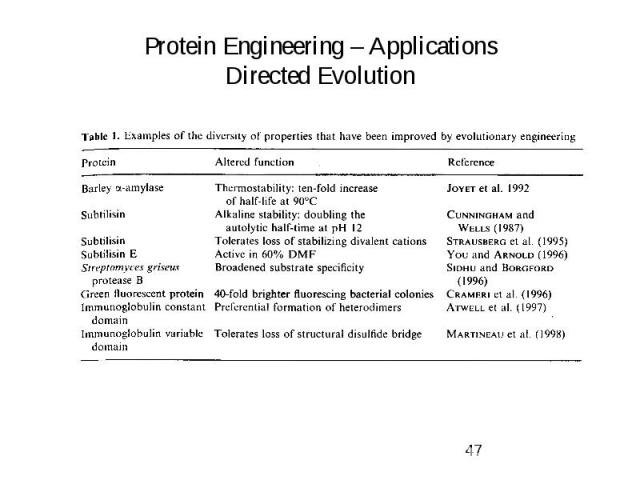
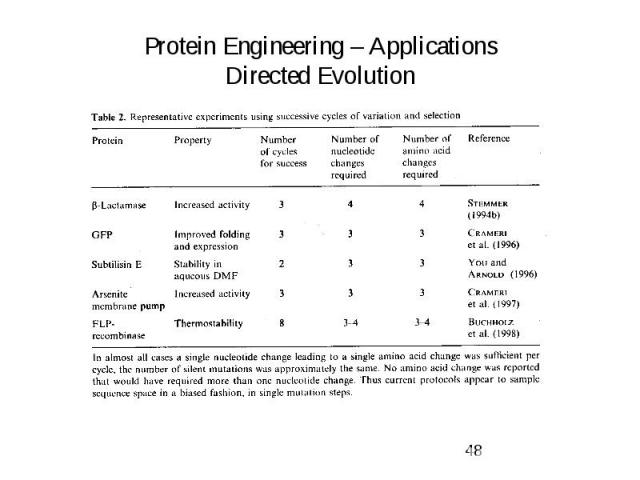
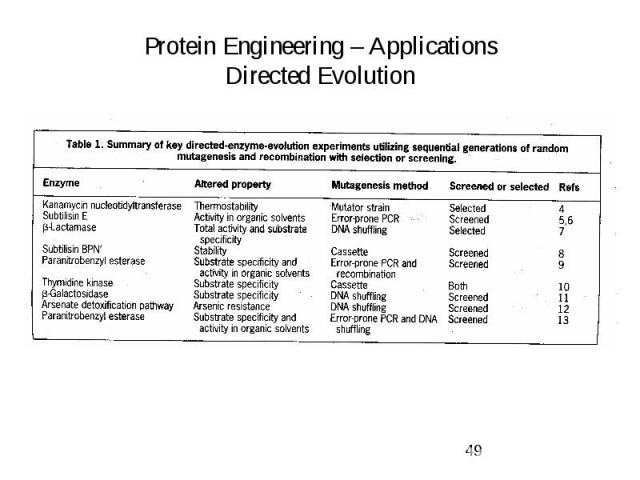
Protein Engineering – Applications Directed Evolution
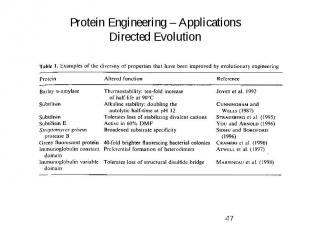
Protein Engineering – Applications Directed Evolution
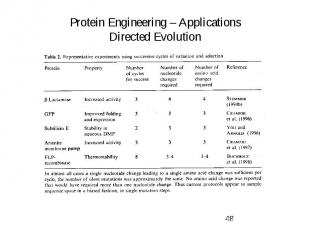
Protein Engineering – Applications Directed Evolution
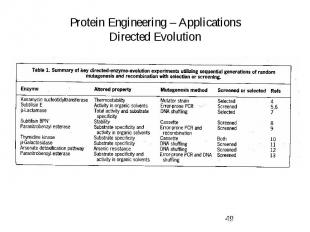
Protein Engineering – Applications Directed Evolution
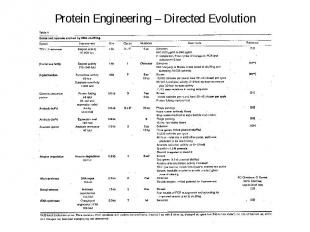
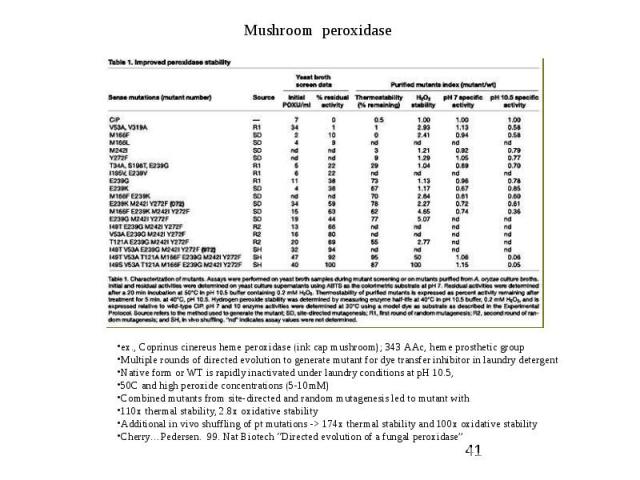
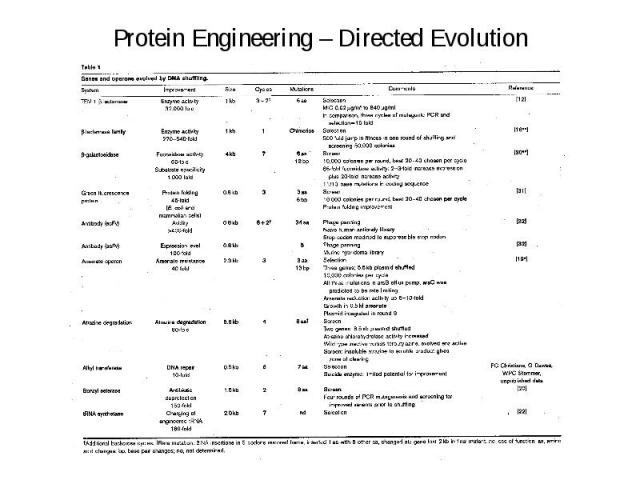
Protein Engineering – Directed Evolution
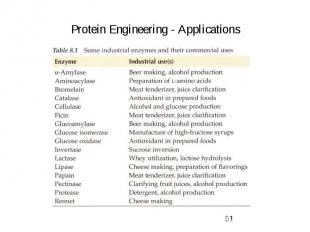
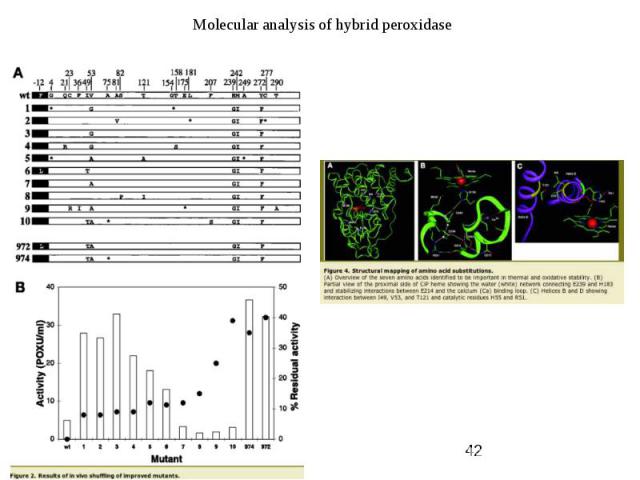
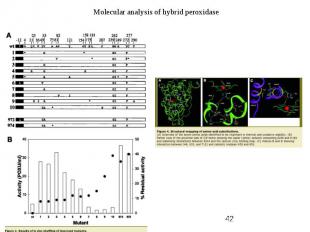
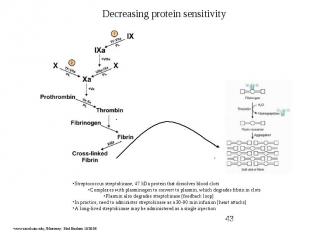
Protein Engineering - Applications